|
 |
- Search
| Korean J Helicobacter Up Gastrointest Res > Volume 23(4); 2023 > Article |
|
Abstract
Objectives
Methods
Results
Notes
Availability of Data and Material
The datasets generated or analyzed during the study are available from the corresponding author on reasonable request.
Conflicts of Interest
Soo-Jeong Cho, a contributing editor of The Korean Journal of Helicobacter and Upper Gastrointestinal Research, was not involved in the editorial evaluation or decision to publish this article. All remaining authors have declared no conflicts of interest.
Authors’ Contribution
Conceptualization: Soo-Jeong Cho. Data curation: Bokyung Kim, Hyunsoo Chung, Sang Gyun Kim, Soo-Jeong Cho. Formal analysis: Gihong Park. Investigation: Gihong Park. Methodology: Sang Gyun Kim, Soo-Jeong Cho. Project administration: Sang Gyun Kim, Soo-Jeong Cho. Resources: Soo-Jeong Cho. Software: Gihong Park. Supervision: Soo-Jeong Cho. Validation: Soo-Jeong Cho. Visualization: Gihong Park. Writing—original draft: Gihong Park. Writing—review & editing: all authors. Approval of final manuscript: all authors.
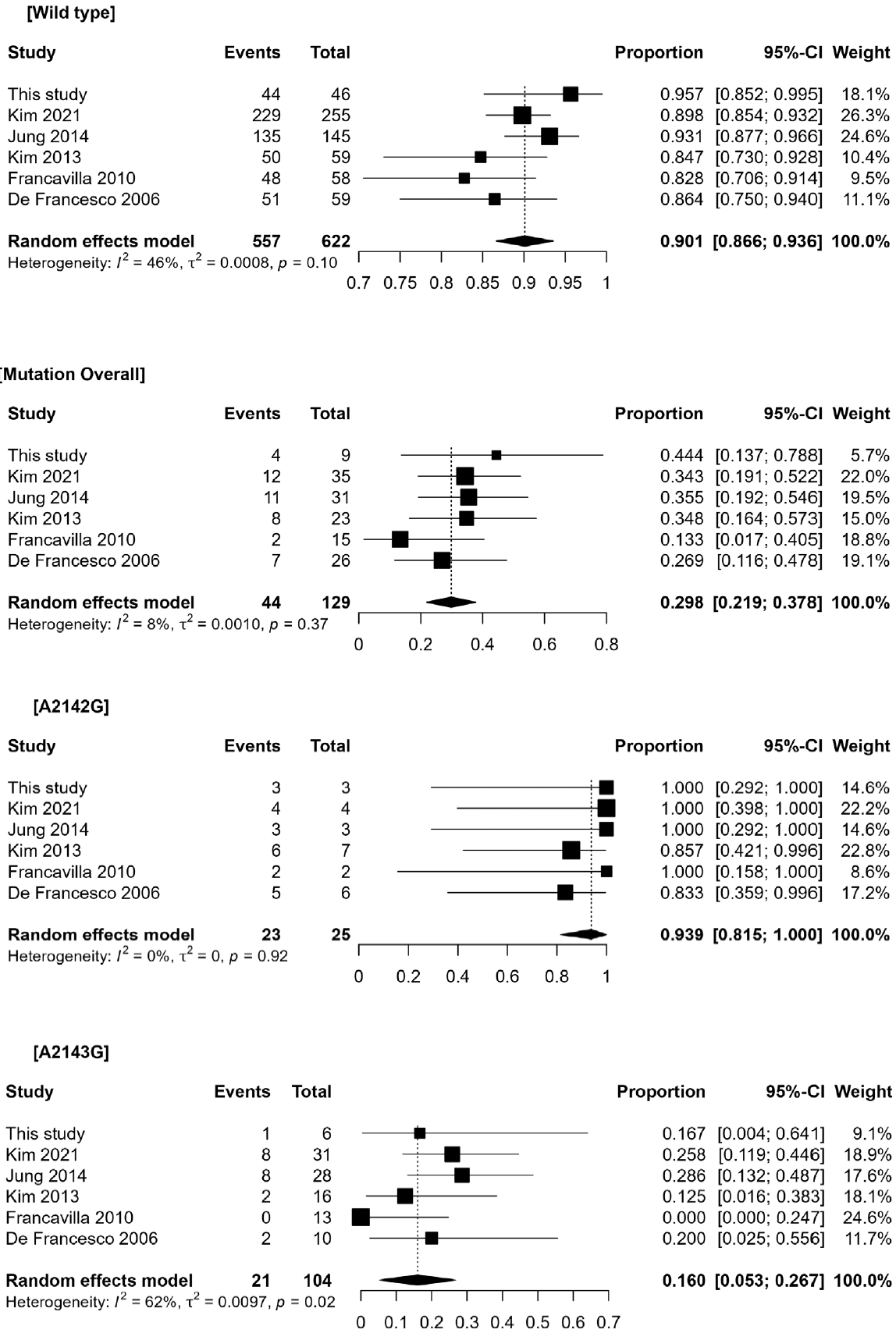
Fig. 1.
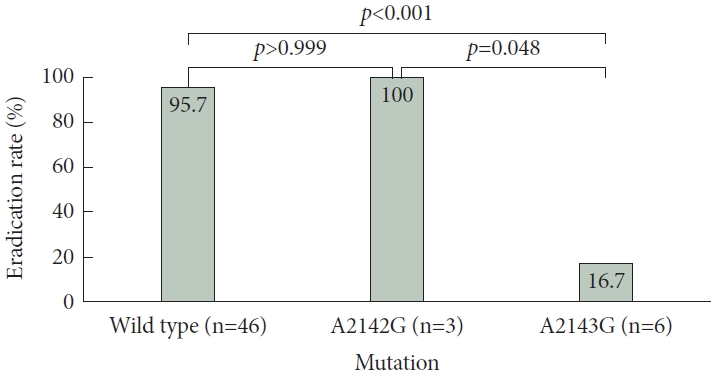
Table 1.
| Characteristics | Wild type (n=46) | A2142G mutation (n=3) | A2143G mutation (n=6) | p-value* |
|---|---|---|---|---|
| Sex | 0.613 | |||
| Male | 24 (52.2) | 2 (66.7) | 5 (83.3) | |
| Female | 22 (47.8) | 1 (33.3) | 1 (16.7) | |
| Age (yr) | 65.0 (60.5–73.5) | 71.0 (59.5–74.0) | 64.0 (62.0–77.0) | 0.834 |
| Hypertension | 26 (56.5) | 2 (66.7) | 3 (50.0) | >0.999 |
| Diabetes | 10 (21.7) | 1 (33.3) | 2 (33.3) | 0.412 |
| Smoking | 13 (28.3) | 1 (33.3) | 3 (50.0) | 0.395 |
| Drinking | 16 (34.8) | 3 (100.0) | 2 (33.3) | 0.061 |
Table 2.
| Studies | Eradication regimen | Wild type | Mutation overall | A2142G mutation | A2143G mutation or double |
|---|---|---|---|---|---|
| This study | Esomeprazole 40 mg | 44/46 (95.6)*† | 4/9 (44.4) | 3/3 (100.0)*‡ | 1/6 (16.7)†‡ |
| Amoxicillin 1 g | |||||
| Clarithromycin 500 mg | |||||
| Twice daily for 10 days | |||||
| Kim et al. [18] (2021) | (Lansoprazole 30 mg or Pantoprazole 40 mg or Rabeprazole 20 mg) | 229/255 (89.8)*† | 12/35 (34.3) | 4/4 (100.0)*‡ | 8/31 (25.8)†‡ |
| Amoxicillin 1 g | |||||
| Clarithromycin 500 mg | |||||
| Twice daily for 7 days | |||||
| Jung et al. [19] (2014) | Standard dose PPI | 135/145 (93.1)*† | 11/31 (35.5) | 3/3 (100.0)*‡ | 8/28 (28.5)†‡ |
| Amoxicillin 1 g | |||||
| Clarithromycin 500 mg | |||||
| Twice daily for 7 days | |||||
| Kim et al. [17] (2013) | Lansoprazole 30 mg | 50/59 (84.7)*† | 8/23 (34.8) | 6/7 (85.7)*‡ | 2/16 (12.5)†‡ |
| Amoxicillin 1g | |||||
| Clarithromycin 500 mg | |||||
| Twice daily for 7 days | |||||
| Francavilla et al. [16] (2010) | Omeprazole (1 mg/kg/day) Amoxicillin (50 mg/kg/day) Clarithromycin (15 mg/kg/day) for 7 days | 48/58 (82.8)*† | 2/15 (13.3) | 2/2 (100.0)*‡ | 0/13 (0.0)†‡ |
| De Francesco et al. [15] (2006) (2006) | Rabeprazole 20 mg | 51/59 (86.4)*† | 7/16 (43.8) | 5/6 (83.3)*‡ | 2/10 (20.0)†‡ |
| Amoxicillin 1g | |||||
| Clarithromycin 500 mg | |||||
| Twice daily for 7 days | |||||
| Overall | 557/622 (89.5) | 44/129 (34.1) | 23/25 (92.0) | 21/104 (20.2) |
* No statistically significant difference in the eradication rate between the wild-type group and the A2142G group (p>0.9);
† The A2143G group demonstrated a significantly lower eradication rate compared to the wild-type group (p<0.05);
Table 3.
REFERENCES
-
METRICS

-
- 1 Crossref
- 1,498 View
- 48 Download
- Related articles in Korean J Helicobacter Up Gastrointest Res
-
What Is the Optimal Drug Regimen for Helicobacter pylori Eradication Therapy?2024 June;24(2)
Antibiotic Resistance and Helicobacter pylori Eradication Therapy2023 September;23(3)